Excitatory Glutamate receptors (dAMPA and NMDA)
This exercise will demonstrate the dAMAP and NMDA glutamate receptors in a single compartment model. Bring up the simulation by typing:
spike both.hoc -
This will bring up two Neuron windows. Close the “NEURON Main Panel”
and retrieve the session file ‘iclamp_glut.ses’ from the “Print & File Window Manager” selecting session/retrieve with the left mouse button

Lots of windows come up after you press “Retrieve from file”!!!
We study the receptor mechanisms here by constructing a single compartment model and inserting the receptors. Within the hoc- file the later is accomplished by the code:
objectvar chan
soma chan=new NMDAR(.5)
objectvar chanN1
soma chanN1 = new dAMPA_R(.50)
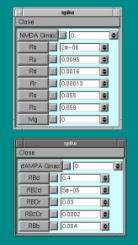
Some parameters of the receptor are controlled by the Neuron tables on the right. We are now concerned with 3 of these values-
NMDA Gmax, dAMPA Gmax and Mg.
Four conditions may be looked at:
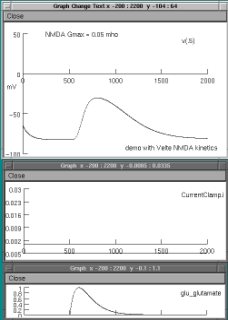
1. NMDA receptor alone: set NMDA Gmax to 0.05 mohs
2. NMDA with Magnesium: as above and set Mg = 0.5 mM
3. DAMPA alone - dAMPA Gmax = 0.05 mohs (don’t forget to set NMDA Gmax = 0)
4 NMDA (with Mg) and dAMPA simultaneously.
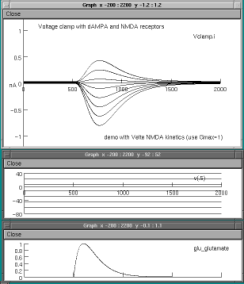
We are running a current-clamp in the compartment and will be observing the voltage excursion there to an application of glutamate to the respective mechanisms. The glutamate concentration over time is determined by an alpha-function trajectory. The current clamp is on for the entire 2000 ms. expressed in the graphs.Shown below is a single clamp record for a holding current of -.005 nA and glutamate applied at 500ms.The top graph shows the voltage;
the middle the command current of the clamp and the bottom the glutamate concentration ,
all over time in ms.
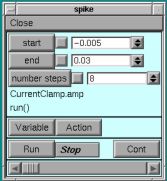
The current clamp protocol used ranges from -.005 to 0.03 nA in 8 steps.
Press “Run” with the left-mouse button to start the simulation.
Before running the clamp experiment, remember to choose “Keep Lines” in the top two graphs using the right-mouse button.
The voltage clamp works in a similar manner. Quit Neuron and bring it up again as before-
spike both.hoc -
but this time retrieve the session file ‘vclamp_glut’
Here we are observing the current excursion rather than voltage. For this condition use the value of 1.0 for NMDA Gmax and dAMPA Gmax.
Run the same 4 conditions that were used for current clamp.
Shown below is a voltage-clamp with 8 steps running from -80 to 40 mV with the NMDA receptor alone.